Neanderthal News - 2020: Part I: the Superarchaics

"The story of human evolution is full of ancient trysts," writes Ann Gibbons in Science. "Genes from fossils have shown that the ancestors of many living people mated with Neanderthals and with Denisovans, a mysterious group of extinct humans who lived in Asia. Now, a flurry of papers suggests the ancestors of all three groups mixed at least twice with even older “ghost” lineages of unknown extinct hominins. One candidate partner: Homo erectus (left, artist's rendition; Yale University), an early human who left Africa by 1.8 million years ago, spread around the world, and could have mated with later waves of human ancestors.
The new genomic studies rely on complex models of inheritance and population mixing, and they have many uncertainties, not least the precise identities of our ancestors’ strange bedfellows and when and where the encounters took place. But, taken together, they build a strong case that even before modern humans left Africa, it was not uncommon for different human ancestors to meet and mate. “It’s now clear that interbreeding between different groups of humans goes all the way back,” says computational biologist Murray Cox of Massey University of New Zealand, Turitea, who was not involved in the new studies.
The gold standard for detecting interbreeding with archaic humans is to sequence ancient DNA from fossils of the archaic group, then look for traces of it in modern genomes. Researchers have done just that with Neanderthal and Denisovan genomes up to 200,000 years old from Eurasia. But no one has been able to extract full genomes from more ancient human ancestors. So population geneticists have developed statistical tools to find unusually ancient DNA in genomes of living people. After almost a decade of tantalizing but unproven sightings, several teams now seem to be converging on at least two distinct episodes of very ancient interbreeding.
In Science Advances today, Alan Rogers, a population geneticist at the University of Utah, Salt Lake City, and his team identified variations at matching sites in the genomes of different human populations, including Europeans, Asians, Neanderthals, and Denisovans. The team tested eight scenarios of how genes are distributed before and after mixing with another group, to see which scenario best simulated the observed patterns. They conclude that the ancestors of Neanderthals and Denisovans—whom they call Neandersovans—interbred with a “super-archaic” population that separated from other humans about 2 million years ago. Likely candidates include early members of our genus, such as H. erectus or one of its contemporaries. The mixing likely happened outside of Africa, because that’s where both Neanderthals and Denisovans emerged, and it could have taken place at least 600,000 years ago.
“I think the super-archaics were in the first wave of hominids who left Africa,” Rogers says. “They stayed in Eurasia, largely isolated from Africans, until 700,000 years ago when Neandersovans left Africa and interbred with them."
Ancestors of Neanderthals and Denisovans interbred with ‘Superarchaic’ Hominin
“We’ve never known about this episode of interbreeding and we’ve never been able to estimate the size of the superarchaic population,” says Professor Alan Rogers in Science Advances.
“We’re just shedding light on an interval on human evolutionary history that was previously completely dark.”
Professor Rogers and colleagues studied the ways in which mutations are shared among modern Africans and Europeans, and ancient Neanderthals and Denisovans (...)
The superarchaic and Neanderthal-Denisovan ancestor populations were more distantly related than any other pair of human populations previously known to interbreed. For example, modern humans and Neanderthals had been separated for about 750,000 years when they interbred.The superarchaics and Neanderthal-Denisovan ancestors were separated for well over a million years.
“These findings about the timing at which interbreeding happened in the human lineage is telling something about how long it takes for reproductive isolation to evolve,” Professor Rogers said.
The researchers used other clues in the genomes to estimate when the ancient human populations separated and their effective population size. They estimated the superarchaics separated into its own species about 2 million years ago. This agrees with human fossil evidence in Eurasia that is 1.85 million years old.
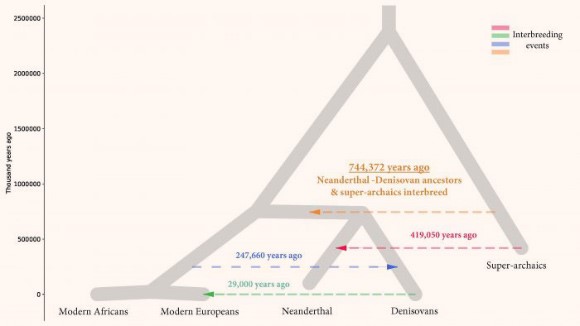
An evolutionary tree including four proposed episodes of gene flow; the previously unknown event 744,372 years ago (orange) suggests interbreeding occurred between superarchaics and Neanderthal-Denisovan ancestors in Eurasia. Image credit: Rogers et al, doi: 10.1126/sciadv.aay5483.
The scientists also proposed there were three waves of human migration into Eurasia.
The first was 2 million years ago when the superarchaics migrated into Eurasia and expanded into a large population.
Then 700,000 years ago, Neanderthal-Denisovan ancestors migrated into Eurasia and quickly interbred with the descendants of the superarchaics.
Finally, modern humans expanded to Eurasia 50,000 years ago where we know they interbred with other ancient humans, including with the Neanderthals.
“I’ve been working for the last couple of years on this different way of analyzing genetic data to find out about history,” Professor Rogers said.
“It’s just gratifying that you come up with a different way of looking at the data and you end up discovering things that people haven’t been able to see with other methods.”
Original article in sci-news.com
Denisovans interbred with archaic hominin
Independently from the University of Utah study, Cornell University researchers Melissa Hubisz and Amy Williams and Adam Siepel of Cold Spring Harbor Laboratory developed a new algorithm for analyzing genomes that can identify segments of DNA that came from other species, even if that gene flow occurred thousands of years ago and came from an unknown source.
The scientists used the algorithm, named ARGweaver-D, to look at genomes from two Neanderthals, a Denisovan and two African humans.
They found evidence that 3% of the Neanderthal genome came from ancient humans, and estimate that the interbreeding occurred between 200,000 and 300,000 years ago.
Furthermore, 1% of the Denisovan genome likely came from an unknown and more distant relative, possibly Homo erectus, and about 15% of these archaic regions were, in turn, introgressed into modern humans and continue to exist in the genomes of people alive today.
The new study has been published in PLos Genetics.
Ann Gibbons points out that new results from other researchers show modern human ancestors mixed with super-archaic groups more recently, in Africa. Population geneticist Sriram Sankararaman and his student Arun Durvasula at the University of California (UC), Los Angeles, identified signs of a separate, more recent episode of mixing. The researchers analyzed the genomes of 405 people from four subpopulations in West Africa that were included in the 1000 Genomes Project, a catalog of genomes from around the world. They found numerous gene variants not seen in Neanderthals or Denisovans and concluded that the best explanation was that the variants came from an archaic, extinct human.
This ghost species may have been late H. erectus, H. heidelbergensis, or a close relative. One or more late-surviving members of this ancient group met and mated with the ancestors of living Africans sometime in the past 124,000 years, the modern genomes suggest.
West Africans carry DNA from archaic hominin
Enrico de Lazaro reports in Science News on the Sankararaman-Durvasula study which "compared Neanderthal and Denisovan DNA with genomes of 405 individuals from West Africa.
The scientists focused on four contemporary West African populations: Yoruba from Ibadan, Esan from Nigeria, Mende from Sierra Leone, and Gambian.
They found differences that could be best explained by introgression (introgression is the process by which members of two populations mate, and the resulting hybrid individuals then breed with members of the parent populations) by an unknown archaic hominin whose ancestors split off from the human family tree before Neanderthals.
The data suggest this introgression may have happened relatively recently, or it may have involved multiple populations of archaic human, hinting at complex and long-lived interactions between anatomically modern humans and various populations of archaic hominins.
“Combining our results across the West African populations, we estimate that the archaic population split from the ancestor of Neanderthals and modern humans 360,000 years to 1.02 million years ago, and subsequently introgressed into the ancestors of present-day Africans 0-124,000 years ago, contributing 2 to 19% of their ancestry,” the authors said.
Dr. Durvasula and Dr. Sankararaman also examined the frequencies of archaic DNA segments to investigate whether natural selection could have shaped the distribution of archaic genetic variants.
“We found 33 loci with an archaic segment frequency of over 50% in Yoruba and 37 loci in Mende,” they said.
“Some of these genes are at high frequency across both Yoruba and Yoruba, including NF1 (a tumor suppressor gene), MTFR2 (a gene involved with mitochondrial aerobic respiration in the testis), HSD17B2 (a gene involved with hormone regulation), KCNIP4 (a gene involved with potassium channels), and TRPS1 TRPS1 (a gene associated with trichorhinophalangeal syndrome).”
“Three of these genes have been found in previous scans for positive selection in Yoruba: NF1, KCNIP4, and TRPS1. On the other hand, we do not find elevated frequencies at MUC7, a gene previously found to harbor signatures of archaic introgression.”
The team calls for more analysis of modern and ancient African genomes to reveal the nature of this complex history.
“The signals of introgression in the West African populations that we have analyzed raise questions regarding the identity of the archaic hominin and its interactions with the modern human populations in Africa,” the researchers said.
“A detailed understanding of archaic introgression and its role in adapting to diverse environmental conditions will require analysis of genomes from extant and ancient genomes across the geographic range of Africa.”
The results were published in the journal Science Advances. Arun Durvasula & Sriram Sankararaman. 2020. Recovering signals of ghost archaic introgression in African populations. Science Advances 6 (7): eaax5097; doi: 10.1126/sciadv.aax5097
Enrico de Lazaro in sci-news.com
Youtube Video of Sriram Sankararaman
Another paper reported Neanderthal DNA in living Africans, likely from migrations back to Africa by early Europeans who bore Neanderthal DNA. Sankararaman thinks some of the archaic DNA he detects in Africans may be from Neanderthals, but most is from the older ghost species. “I think the true picture is a combination of both an archaic population unrelated to Neanderthals as well as Neanderthal-related ancestry,” he says.
African individuals carry more Neanderthal DNA than previously thought
Using a novel method for detecting Neanderthal ancestry in modern humans, a team of U.S. researchers has demonstrated that remnants of Neanderthal genomes survive in every modern human population studied to date. They have found that modern African individuals have more Neanderthal DNA than previously thought; this can be explained by genuine Neanderthal ancestry due to migrations back to Africa, predominately from ancestral Europeans.
“Our study is significant because it provides important new insights into human history and patterns of Neanderthal ancestry in globally diverse populations,” said senior author Dr. Joshua Akey, a scientist at Princeton University.
“Our results refine catalogs of genomic regions where Neanderthal sequence was deleterious and advantageous and demonstrate that remnants of Neanderthal genomes survive in every modern human population studied to date.”
Dr. Akey and his colleagues from Princeton University, the University of Washington, Seattle, and Microsoft developed a new computational method for detecting Neanderthal ancestry in the human genome.
Called IBDmix, their method enabled them for the first time to search for Neanderthal ancestry in African populations as well as non-African ones.
“This is the first time we can detect the actual signal of Neanderthal ancestry in Africans. And it surprisingly showed a higher level than we previously thought,” said co-author Dr. Lu Chen, a postdoctoral research associate in the Lewis-Sigler Institute for Integrative Genomics at Princeton University.
The IBDmix method draws its name from the genetic principle ‘identity by descent’ (IBD), in which a section of DNA in two individuals is identical because those individuals once shared a common ancestor.
The length of the IBD segment depends on how long ago those individuals shared a common ancestor.
For example, siblings share long IBD segments because their shared ancestor (a parent) is only one generation removed. Alternatively, fourth cousins share shorter IBD segments because their shared ancestor (a third-great grandparent) is several generations removed.
The researchers applied IBDmix to 2,504 modern individuals from the 1,000 Genomes Project, which represents geographically diverse populations, and used the Altai Neanderthal reference to identify Neanderthal sequence in these individuals.
They robustly identified regions of Neanderthal ancestry in Africans for the first time, identifying on average 17 megabases (Mb) of Neanderthal sequence per individual in the African samples analyzed (which corresponds to approximately 0.3% of the genome), compared with less than one megabase reported in previous studies.
More than 94% of the Neanderthal sequence identified in African samples was shared with non-Africans.
The scientists also observed levels of Neanderthal ancestry in Europeans (51 Mb/individual), East Asians (55 Mb/individual), and South Asians (55 Mb/individual) that were surprisingly similar to each other.
Strikingly, East Asians had only 8% more Neanderthal ancestry compared to Europeans, in contrast to previous reports of 20%.
“This suggests that most of the Neanderthal ancestry that individuals have today can be traced back to a common hybridization event involving the population ancestral to all non-Africans, occurring shortly after the Out-of-Africa dispersal,” Dr. Akeysaid.
To explore potential explanations for the unexpectedly high Neanderthal ancestry in Africans, the researchers then compared the actual data to simulated genotype data derived from different demographic models.
They found that Africans exclusively share 7.2% of Neanderthal sequence with Europeans, compared with only 2% with East Asians.
Simulations showed that low levels of back-migration persisting over the past 20,000 years can replicate features of the data and could therefore be a possible explanation for the observed levels of ancestry among different modern populations.
But gene flow went in both directions. The data also suggest that there was a dispersal of modern humans out of Africa approximately 200,000 years ago, and this group hybridized with Neanderthals, introducing modern human DNA into the genomes of Neanderthals.
“Both out-of-Africa and into-Africa dispersals must be accounted for when interpreting global patterns of genomic variation,” the study authors said.
A paper on the findings was published in the journal Cell. (Lu Chen et al. Identifying and Interpreting Apparent Neanderthal Ancestry in African Individuals. Cell, published online January 30, 2020; doi: 10.1016)
Original article in sci-news.com
Ann Gibbons lists yet another study, of December 2019, which "found hints of an extinct ghost population in living Africans, although it was silent on the identity of the ghosts and when they bred with our ancestors. Population geneticist Jeff Wall at UC San Francisco and colleagues analyzed 1667 genomes from diverse populations in the GenomeAsia 100K consortium. They reported the strongest ghost signal in the Khoisan people and in Central African hunter-gatherers formerly known as pygmies.
But Wall and others warn their methods cannot rule out that the “ghosts” could be one or several groups of modern humans in Africa that were separated from other moderns for so long that their genes looked “archaic” when the groups finally came together again and mixed. “Our understanding of African population history, in particular, is so far behind,” says Joshua Akey of Princeton University.
Even if they differ on particulars, the studies emphasize that long after new lineages of humans emerge, they still can mix with others quite different from themselves. Other species, such as cave bears and mammoths, show the same pattern of divergence and later mixing, says population geneticist Pontus Skoglund of the Francis Crick Institute in London. “We are losing the idea that separation between populations is simple with instant isolation.” Such mating between long isolated groups may quickly introduce valuable new genes. For example, some of the archaic alleles Sankararaman spotted in Africans were in genes that suppress tumors and regulate hormones.
Today, H. sapiens doesn’t have the possibility of quickly grabbing a load of diversity by mating with another group: For perhaps the first time in our history, we’re the only humans on the planet. It’s another reason to miss our extinct cousins, says population geneticist Carina Schlebusch of Uppsala University.
“To have such a large densely spread species with … so little genetic diversity … is a dangerous situation,” she says.
(Ann Gibbons, Science.)

